
May 6, 2020 — Following Scott Marshall’s announcement last week about the QuartzBio team’s COVID-19-Related Data Aggregation Initiative, we wanted to share additional context behind the scope of our efforts. Our initial focus is creating a valuable and thoughtfully constructed data ecosystem, which will result in the ability to draw meaningful context. This is nucleated with published data from critical SARS-CoV-2 profiling experiments, that compare infection of cell line and animal models with novel coronavirus to other viruses like SARS-CoV, MERS-CoV and H1N1.
While we expect this to rapidly evolve given the dramatic pace of global research—today limited -omics (e.g,. RNAseq) data from COVID-19 positive clinical samples (at scale) are available. However, we can use rich clinical profiling data from other published sources like GTEx and TCGA as a starting point for translational research. For example, many groups are identifying correlative relationships between the SARS-CoV-2 receptor ACE2, with other molecules (e.g., inflammatory mediators). Efforts such as these can help us systematically evaluate potential therapeutic signaling pathways by leveraging existing, large-scale resources that afford opportunities not only to stratify by molecular profile, but also consider comorbidities, or age and other factors.
The table below is not exhaustive but highlights some of the initial data we are including in our data aggregation effort and the scientific rationale (at present).
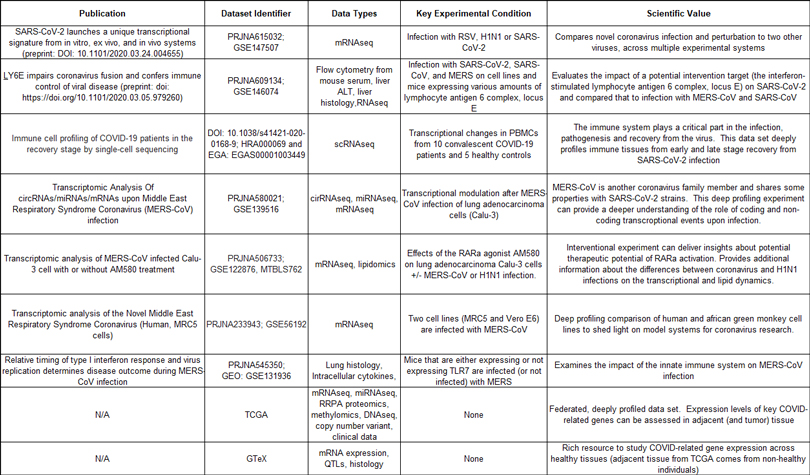
Our data aggregation is an ongoing and dynamic initiative. As relevant data sets come online, or as ongoing research highlights new potential directions, we will continue to add and refine. This includes the potential for expansion beyond our largely molecular first steps: as our effort is phased and collaborative we would welcome suggestions on relevant molecular data sets and / or other types of biological data that should be considered.
In our next post, we’ll share more about the work that goes into integrating these diverse data sources to enable cross-dataset exploration and visualization. In the mean time, we’d love to hear about what other profiling data sets you would find valuable to add to this ecosystem!
Author: Renée Deehan, VP, Computational Biology at QuartzBio, part of Precision for Medicine